Image annotation and bio-image database
Guillaume Gay, CENTURI
June 10 2021
We are here to talk about microscopy image databases. We are not going to talk a lot about the “database” part of that, because a lot has to be said about microscopy and images before that is more important.
Follow this course here:
Why you should care?

Excel is an accounting tool
Course outline
Some history of microscopy techniques
The digital image (data and metadata)
Databases (at last)
Some technique history
Early Microscopes
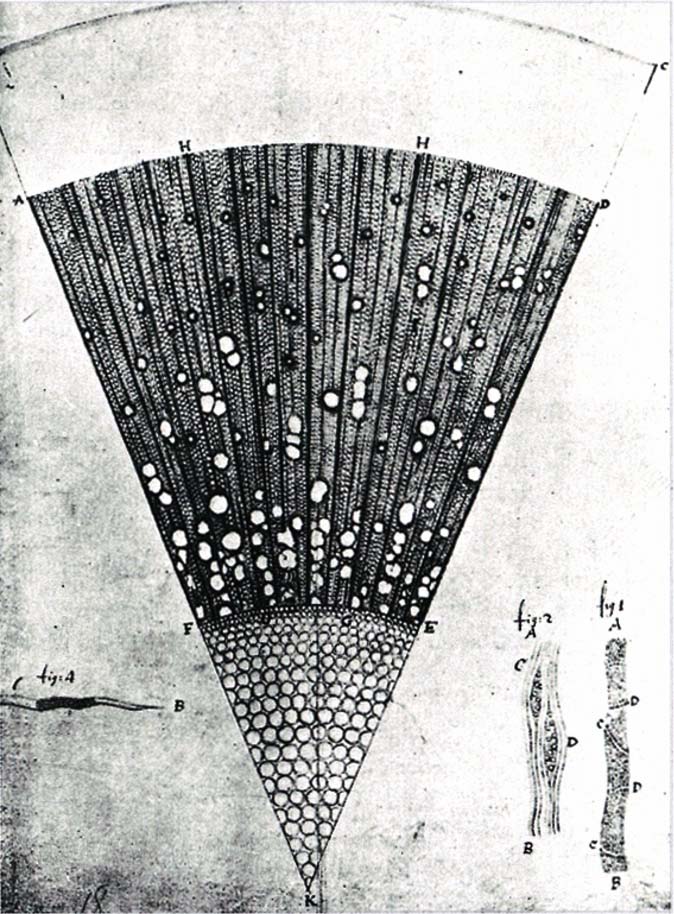
First detector is the eye, data is registered through drawings.
Santiago Ramón y Cajal (1852 - 1934)

Public Domain, Link
The eye & hand are still the best detector in the early XXth century.
First photos
Henry Fox Talbot (1800 - 1877)

See this article
First movies
Haemanthus katherinae (1956!)
Technique evolution
Fluorescence !
- dark field
- multiple colors
- specificity - we observe not only the organism but a precise molecule within the organism.
The confocal microscope
- The detector is a photomuliplier - first time the image from the microscope is a signal
- Only the light emitted at the focal point is recorded.
Here comes the CCD
Some details on how it works here
Photon counting!
The image is a quantitative, digital, signal
From now on, an image is represented by a matrix of pixels
Modern microscopes
The super resolution revolution
Do you know Abbe law?
\[d = \frac {\lambda}{2 n A} \]
The minimum size of a motif - for exemple the distance between two spots, observable under a microscope is limited by the objective numerical aperture and the emission wavelength.
We invented ways to beat that limit!
(can you cite super resolution methods?)
An exemple: STORM

An other: Lattice light sheet
Movie 11 High Resolution from HHMI NEWS on Vimeo.
Sreens and plates
Multiple wells under a microscope on a moving stage

Conclusion
Image aquisition methods have always been immediatly applied to microscopy
The eye was surpassed only recently
The image became digital only 20 years ago!
The digital image
What is important to know?
Data and metadata
Can you cite image formats?
TIFF is the norm
TIFF is for Tagged Interchange File Format
A TIFF is a structured file with a header before the data:

We have tags to store metadata !

What an 8 by 8 pixel file looks like:

00000000: 4949 2a00 0800 0000 0e00 0001 0400 0100 II*.............
00000010: 0000 0800 0000 0101 0400 0100 0000 0800 ................
00000020: 0000 0201 0300 0100 0000 0800 0000 0301 ................
00000030: 0300 0100 0000 0100 0000 0601 0300 0100 ................
00000040: 0000 0100 0000 0e01 0200 1200 0000 b600 ................
00000050: 0000 1101 0400 0100 0000 3001 0000 1501 ..........0.....
00000060: 0300 0100 0000 0100 0000 1601 0400 0100 ................
00000070: 0000 0800 0000 1701 0400 0100 0000 4000 ..............@.
00000080: 0000 1a01 0500 0100 0000 0801 0000 1b01 ................
00000090: 0500 0100 0000 1001 0000 2801 0300 0100 ..........(.....
000000a0: 0000 0100 0000 3101 0200 0c00 0000 1801 ......1.........
000000b0: 0000 0000 0000 7b22 7368 6170 6522 3a20 ......{"shape":
000000c0: 5b38 2c20 385d 7d00 0000 0000 0000 0000 [8, 8]}.........
000000d0: 0000 0000 0000 0000 0000 0000 0000 0000 ................
000000e0: 0000 0000 0000 0000 0000 0000 0000 0000 ................
000000f0: 0000 0000 0000 0000 0000 0000 0000 0000 ................
00000100: 0000 0000 0000 0000 0100 0000 0100 0000 ................
00000110: 0100 0000 0100 0000 7469 6666 6669 6c65 ........tifffile
00000120: 2e70 7900 0000 0000 0000 0000 0000 0000 .py.............
00000130: 0101 0101 0101 0101 0101 0101 0101 0101 ................
00000140: 0101 0101 0101 0101 0101 0101 0101 0101 ................
00000150: 0101 0101 0101 0101 0101 0101 0101 0101 ................
00000160: 0101 0101 0101 0101 0101 0101 0101 0101 ................A sad story
- In the 90’s - 2000’s, MetaMorph software dominates the industry, has its own ‘format’
- Eventually, constructors build their own software, try to impose it, how?
\(\Rightarrow\) Lots of incompatible & proprietary formats
OME to the rescue
«It is possible to interpret images only if we know the context in which they were acquired»
The OME-TIFF Format
We can put “things” in the TIFF header - so why not all the metadata we can think off?
This became a standard
The whole schema
Is available here
Look for the important points you thought of.
What do you think of XML?
In a file
<Image ID="Image:0" Name="Excy2_4.6.+12.lif [Excy2 4.6 - Phall CD24 Org 2]">
<AcquisitionDate>2016-05-20T13:08:29
</AcquisitionDate>
<ImagingEnvironment/>
<Pixels BigEndian="true"
DimensionOrder="XYCZT"
ID="Pixels:0"
Interleaved="false"
PhysicalSizeX="0.4814710371819961"
PhysicalSizeXUnit="µm"
PhysicalSizeY="0.4814710371819961"
PhysicalSizeYUnit="µm"
SignificantBits="8"
SizeC="4"
SizeT="1"
SizeX="512"
SizeY="512"
...See more details here
Limits to OME-XML
What did you say a microscope image was?
The future: how to define flexible, “just general enough” file formats.
Let’s look at ZARR
The other metadata
But there’s more! The organism, the protocol, gene deletion,

Resort to ontologies
Global consortium QUAREP - LiMi
Finally Databases!
One DB system to rule them all: OMERO
- Postgresql based
- Lots of features
The Contender Cytomine (but still using BioFormats!)
Cytomine is oriented towards collaboration after the image is produced.
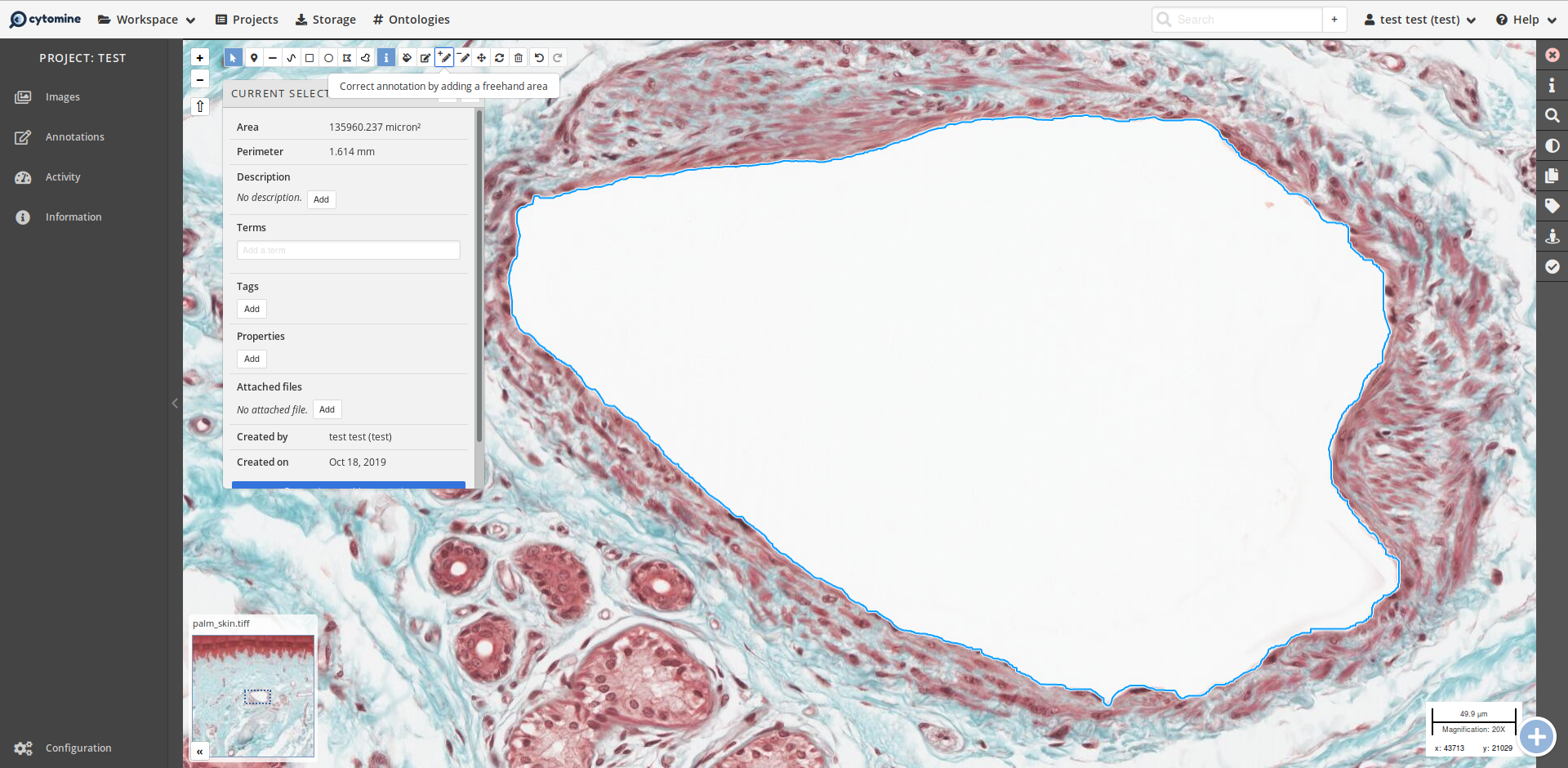
Public microscopy image databases
A word on FAIR
- We need to be able to reuse data
- We must be able to do this automatically
A non exhaustive list:
The Allen Institute
Brain Atlases
The Allen Cell explorer
- Tries to know all the possible states of stem cells
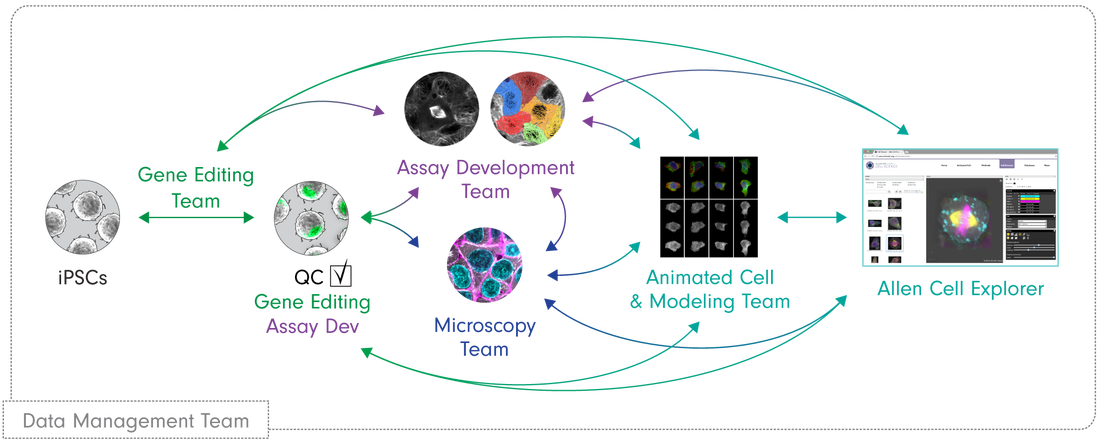
- Created an extensive catalog of cell structures
4D Nucleome
A platform to search, visualize, and download nucleomics data.
Icludes microscopy data
European initiatives
Under the BioImage Archive
The Future ™
- Lots of efforts towards FAIR - Fr, EU, Worldwide infrastructure
- Crossing microscopy with other *omics data
- Setting up standards is very hard
Conclusion
Already a lot of ressources but
- Little actual re-use for now
- Not used everywhere (biologists are still reluctant to share)
- Please annotate your data
- Talk to me if you need omero!